The EuroEPINOMICS live ticker. During the last ten months, the focus of the EuroEPINOMICS consortium on Rare Epilepsy Syndromes (RES) has been the trio analysis in patients with epileptic encephalopathies. The RES subproject also aims at analysing the genetics of rare familial epilepsies. There have been ongoing discussions about the best way to get the analysis started. Last week, we finally “sealed the deal” for whole genome analysis in RES families. The EuroEPINOMICS RES consortium will collaborate with the CoGIE partner Luxembourg Centre for Systems Biomedicine (LCSB), the Institute of Systems Biology, Seattle USA (ISB) and Complete Genomics to analyse whole genome data of families with Rare Epilepsy Syndromes with an expected capacity of 100 genomes. But why exactly are we thinking about trading exomes for whole genomes?
Whole genomes are not just big exomes. If you think that the data analysis of a human genome is simply exome data scaled up by the factor of >100, think again. There are different rules at play in the analysis of whole genome data. First of all, quality issues are actually less of a concern for whole genome data than exome data. The expected mismatch between provided sequence and actual sequence is 1:30,000 or less. Therefore, the argument that a lack of gene findings might be due to lack of coverage in particular gene regions does not apply to Complete Genomics whole genome data. Genomic data can be assumed to be complete, which opens up a completely new horizon of possibilities. Finally, high-throughput genetic data is no longer a screening tool, but an assessment tool for genomic completeness.
Linkage-free linkage with gene findings on the side. With high quality genomic data, the transmission of every single variant in the human genome can be assessed according to predefined criteria. For example, all variants in the human genome can be queried as to whether they are transmitted in an autosomal dominant fashion. This inheritance state analysis was developed at the ISB for family quartets and can be applied to larger families in a modified form. Though computationally challenging it is possible to filter the number of variants in a medium-sized family to a few potentially causal variants. Inheritance state analysis is pretty much linkage-free linkage. Linkage is obsolete, as nothing needs to be linked. We can directly look at each variant in the human genome. We aim to apply methods like this to the EuroEPINOMICS RES families to identify the underlying causative genes.
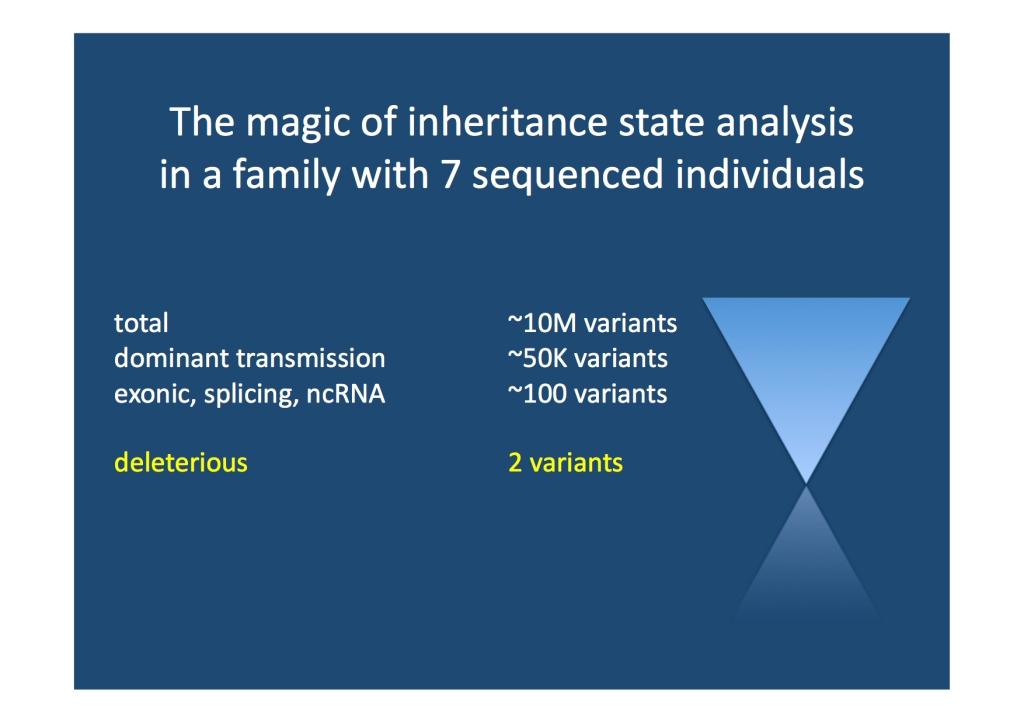
Linkage-free linkage. Inheritance state analysis is capable of zooming in on a relatively small number of variants in a medium sized family. No linkage involved.
Selection of families for WGS. EuroEPINOMICS RES has established working groups for autosomal dominant and recessive epilepsy families. We have contacted all partners already regarding a date for a teleconference and would ask everybody who has not received the initial email to complete the Doodle invite. We probably have capacity for up to 100 genomes, which will probably correspond to 20-30 epilepsy families.
The analysis team. We are very happy that this landmark project of the EuroEPINOMICS consortium is already equipped with a more than capable analysis team. We are looking forward to working with our partners from LCSB and the Family Genomes group at the ISB. On the Luxembourg site, we would like mention Patrick May, Roland Krause and Rudi Balling who have taken on the task of analysing the data. The steels mills and servers at the LCSB are already spinning, ready to crunch some RES genomes.



